Click the links below to jump to the corresponding download page.
Prunus persica
Prunus davidiana
Prunus mira
Prunus ferganensis
Prunus kansiensis
Identification and annotation of TE sequences
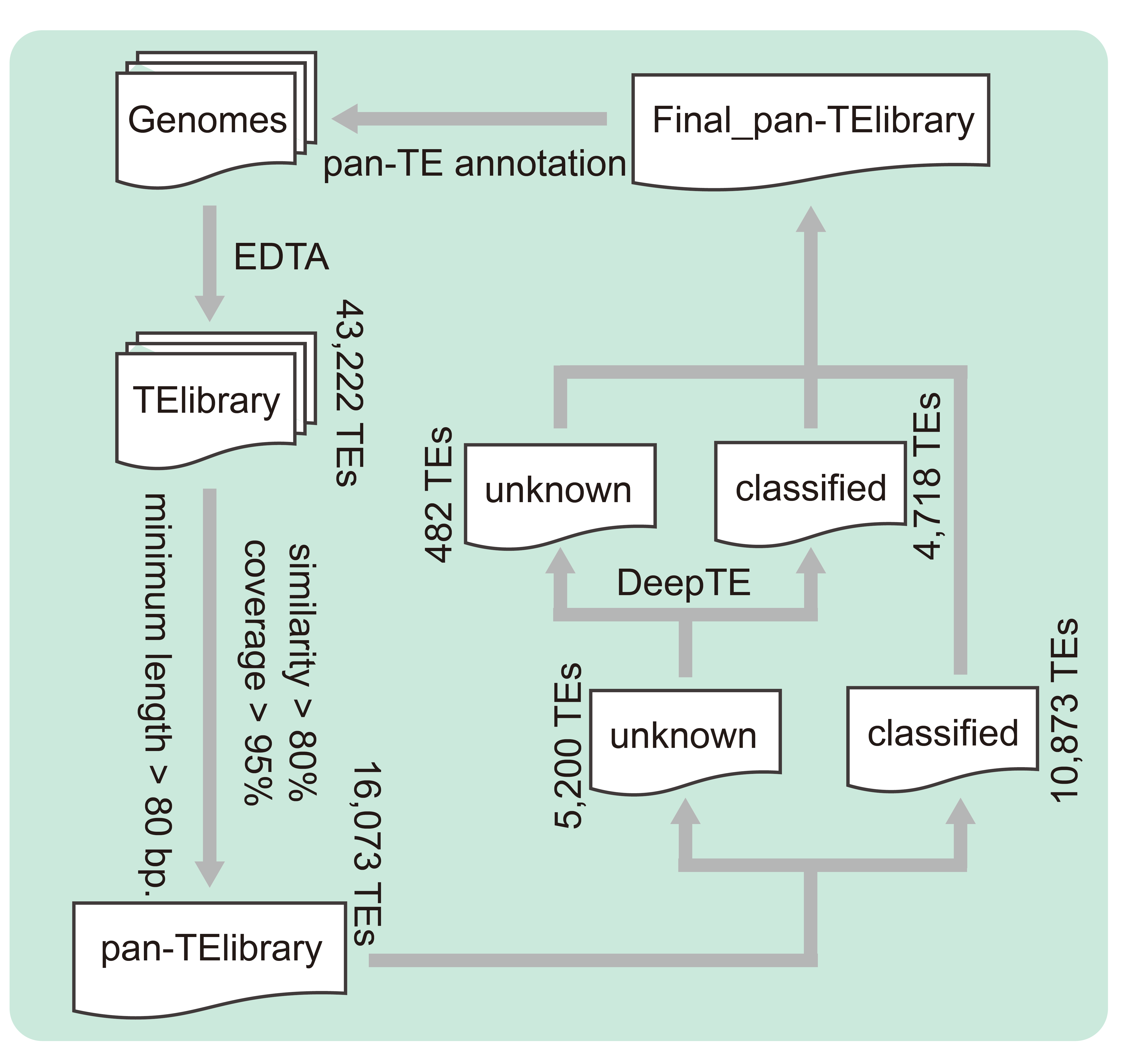
TE libraries were constructed using EDTA without RepeatModeler turned on for structural annotation of each genome. The pan-TE library was constructed using the pan-EDTA pipeline with parameters sequence similarity > 80%, coverage > 95%, and minimum length > 80 bp. DeepTE was used to further annotate the unclassified TE families in pan-TE library. The EDTA pipeline was then reapplied with RepeatModeler turned on to re-annotate transposons for each genome using the pan-TE library as a corrective library. Based on the TE annotation results and genome annotation files, a bed format file was generated using bedtools and the percentage TE content in genomic elements was calculated using the bedtools -coverage command. Considering the high heterozygosity of the peach genome, to avoid false positives, we first extracted the gene sequences and then performed TE annotation using RepeatMasker (https://github.com/Dfam-consortium/RepeatMasker/) with constructed TE libraries.